Section: New Results
Meristem functioning and development
In axis 2 work focuses on the creation of a virtual meristem, at cell resolution, able to integrate the recent results in developmental biology and to simulate the feedback loops between physiology and growth. The approach is subdivided into several sub-areas of research.
Data acquisition and design of meristem models
Participants : Frédéric Boudon, Christophe Godin, Vincent Mirabet, Jan Traas, Grégoire Malandain, Jean-Luc Verdeil.
This research theme is supported by the ATP CIRAD Meristem and the ANR GeneShape and FlowerModel projects.
Studies on plant development require the detailed observation of the tissue structure with cellular resolution. In this context it is important to develop methods that enable us to observe the inner parts of the organs, in order to analyze and simulate their behavior. Here we focus on the apical meristems, that have been extensively studied using live imaging techniques and confocal microscopy. An important limitation of the confocal microscope lies in the data anisotropy. To overcome this limitation, we designed new protocols to achieve an accurate segmentation of the cells. Using these segmentations, a geometrical and topological representation of the meristem is built. Such representations may be used to analyze the meristem structure at cell level, to support the description of gene expression patterns and to initiate and assess virtual meristem simulations.
-
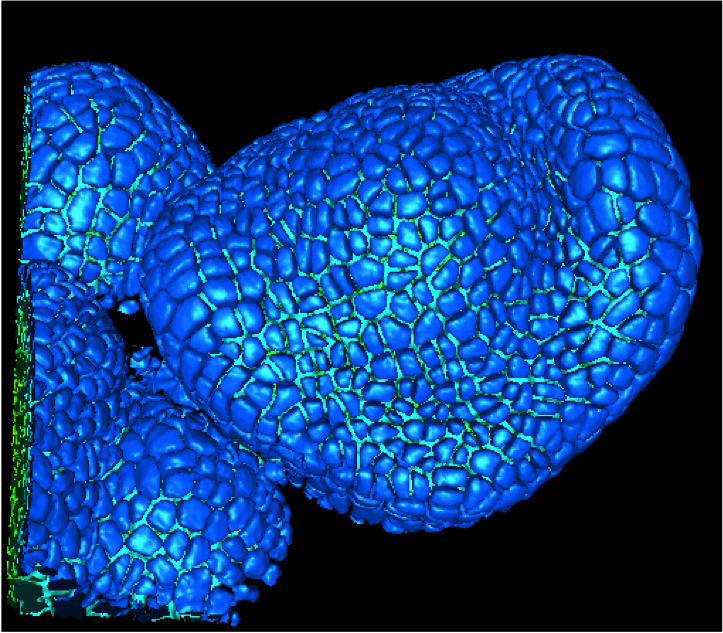
Microscopy image reconstruction and automatic lineage tracking of the growing meristem cells
Participants : Romain Fernandez, Christophe Godin, Grégoire Malandain, Jan Traas, Pradeep Das, Vincent Mirabet.
In previous work [5] , we studied the tracking of meristem cells using time-lapse confocal microscopy acquisition on early stages flowers of Arabidopsis shoot apical meristems. We designed a reconstruction method (MARS, Figure 4 ) and a tracking algorithm (ALT) in order to map the segmentations of the same meristem at different times, based on a network flow representation in order to solve the cell assignment problem. We validated the MARS-ALT pipeline on a four-steps time course of an early stage floral bud. In 2011, we worked to improve the robustness of the MARS-ALT pipeline. The software pipeline was completely re-engineered so as to be easily available in OpenAlea with documentation. We also designed new tests to improve the method and get high-quality results on different types of organisms.
-
Design of 3D virtual maps for specifying gene expression patterns (Jérôme Chopard, Christophe Godin, Jan Traas, Françoise Monéger [ENS Lyon])
This research theme is supported the ANR GeneShape and FlowerModel projects.
To organize the various genetic, physiological, physical, temporal and positional informations, we build a spatialized and dynamic database. This database makes it possible to store all the collected information on a virtual 3D structure representing a typical organ. Each piece of information has to be located spatially and temporally in the database. Tools to visually retrieve and manipulate the information, quantitatively through space and time are being developed. For this, the 3D structure of a typical organ has been created at the different stages of development of the flower bud. This virtual structure contains spatial and temporal information on mean cell numbers, cell size, cell lineages, possible cell polarization (transporters, microtubules), and gene expression patterns. Such 3D virtual map is mainly descriptive. However, like for classical databases, specific tools make it possible to explore the virtual map according to main index keys, in particular spatial and temporal keys. Both a dedicated language and a 3D user interface are being designed to investigate and query the 3D virtual map.
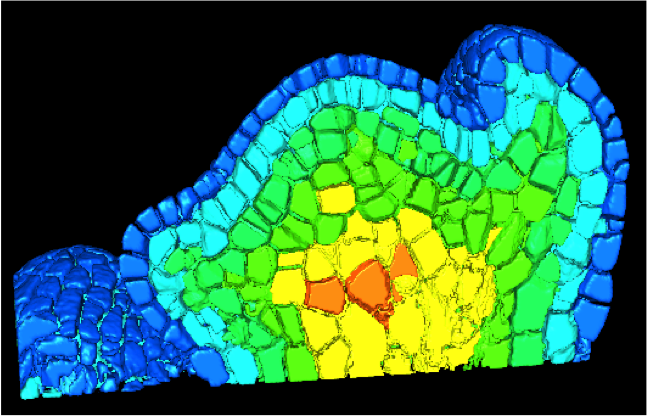
A prototype version of this 3D virtual map is currently being built and is integrated in V-Plants, see figure 5 where a cell-based volumic tissue that can contain different types of information (cell lineage, cell size, cell identity, etc...).
Figure 5. A 3D database represented as a cell-based map that can contain different types of information (cell lineage, cell size, cell identity, ...), from [14] . 
Using this 3D virtual map prototype, along with piecewise-linear models of gene networks, and optimization techniques, our colleagues from ENS/Lyon have been able to develop a model of the gene network regulating sepal polarization in A. thaliana. This model, which is consistent with a very comprehensive literature data set, also include new predictions for certain gene interactions, and will appear next year in Plant Cell .[14]
Shape analysis of meristems
(Jean-Baptiste Durand, Frederic Boudon, Yann Guedon, Francois Mankessi [BURST, DAP], Olivier Monteuuis [BURST, DAP], Jean Luc Verdeil [PHIV, DAP] )
Plants that grow several forms or type of leaves along a shoot, depending on age or shoot length, are called heteroblastic.The influence of heteroblasty on morphological and histocytological characteristics of Acacia mangium shoot apical meristems (SAMs) was assessed comparing materials with mature and juvenile leaf morphology in natural and in vitro conditions. For this we introduced a workflow for characterizing dome shape with few parameters (SAM dome heigth (H), basal diameter (D) and shape factor (S)) and their joint statistical analysis to assess influence of conditions on SAM shape. In particular, a new statistical test is introduced here for multivariate analysis. This is a generalization of univariate ANOVA that takes into account statistical dependencies between the shape parameters. As a result, we found that SAM dome height (H) and basal diameter (D) were highly correlated. The joint analysis revealed that H, D, and shape (S) varied significantly according to the four plant origins investigated, with the higher scores for the outdoor mature source “Mat”. Overall, heteroblasty induced more conspicuous differences of SAM characteristics for the outdoor than for the in vitro materials.
Transport models
Participant : Michael Walker.
This research theme is supported by the ANR GeneShape and ERASysBio+ iSAM projects.
Active transport of the plant hormone auxin has been shown to play a key role in the initiation of organs at the shoot apex, and vein formation in both leaves and the shoot apical meristem. Polar localized membrane proteins of the PIN1 and AUX/LAX family facilitate this transport and observations and models suggest that the coherent organization of these proteins in the L1 layer is responsible for the creation of auxin maxima (surrounded by a depletion zone), which in turn triggers organ initiation close to the meristem center [39] [1] . Furthermore, canalized PIN allocations are thought to play a crucial role in vein formation in the leaf and in the L2.
Previous studies have typically modeled the L1 and L2 with different models to explain their different PIN allocations. For example [25] used the so-called 'up-the-gradient' model in the L1 to recreate the depletion zone around a maximum, and a 'flux-mediated' model to produce the midvein growing through the L2. A more unified approach was that of Stoma et.al. [10] who used the flux-mediated model in both tissues, but with different powers of the auxin flux, i.e. with a linear function in the L1 and a quadratic one in the L2.
We seek a completely unified model with no difference in the PIN allocation model. Our approach is based on inherent differences between the L1 and L2, specifically their dimensionality and the distribution of sources and sinks. We also use a flux-feedback function whose power is intermediate between one and two. For powers close to but slightly higher than one, the diffuse PIN distribution is retained in the L1 but a canalized one is formed in the L2.
We are also comparing two and three dimensional simulations to test the common approximation of three-dimensional tissues being modeled as two-dimensional. We find that source-sink models are well-approximated in two dimensions while sink- and source-driven systems show qualitatively different behavior.
A paper on this unified model is being written. This work is done in the context of the Geneshape and iSAM projects.
Mechanical model
Participants : Jérôme Chopard, Christophe Godin, Frédéric Boudon, Jan Traas, Olivier Hamant [ENS-Lyon] , Arezki boudaoud [ENS-Lyon] .
This research theme is supported by the ANR VirtualFlower and Geneshape projects.
The rigid cell walls that surround plant cells is responsible for their shape. These structures are under constraint due to turgor pressure inside the cell. To study the overall shape of a plant tissue and morphogenesis, its evolution through time, we therefore need a mechanical model of cells. We developed such a model, in which walls are characterized by their mechanical properties like the Young modulus which describes the elasticity of the material. Wall deformation results from forces due to turgor pressure. Growth results from an increase in cell wall synthesis when this deformation is too high. The final shape of the tissue integrates mechanically all the local deformations of each cell.
To model this process, we used a tensorial approach to describe both tissue deformation and stresses . Deformations were decomposed into elementary transformations that can be related to underlying biological processes. However, we showed that the observed deformations does not map directly local growth instructions given by genes and physiology in each cell. Instead, the growth is a two-stage process where genes are specifying by their activity a targeted shape for each cell (or small homogeneous region) and the final cell shape results from the confrontation between this specified shape and the physical constraints imposed by the cell neighbors. Hence the final shape of the tissue results from the integration of all these local rules and constraints at organ level. This work is being described in a paper which will be submitted for publication at the beginning of 2011.
Gene regulatory networks
Modeling gene activities within cells is of primary importance since cell identities correspond to stable combination of gene level activity.
-
The auxin signaling pathway (Etienne Farcot, Yann Guédon, Christophe Godin, Yassin Refahi, Jonathan Legrand, Jan Traas, Teva Vernoux)
The auxin signalling network involves about 50 potentially interacting factors. We applied a graph clustering method [16] that relies on 0/1 interactions between factors deduced from yeast two-hybrid (Y2H) data. The Y2H analysis involves two independent tests (X-gal and HIS3 tests). Each possible interaction was tested in the two possible configurations, where each protein was alternatively the bait and the prey protein. A binary interaction is thus a summary of the four outputs of the X-gal and HIS3 tests. In order to limit the loss of information, we designed a standardization procedure to summarize the outputs of the X-gal and HIS3 tests as a distance defined on a continuous scale. This opens the possibility to studies the influence of phylogenetic distances between factors on their interactions using an extension of the mixture model for random graphs that incorporate explanatory variables. This work is the object of a collaboration with Jean-Benoist Leger and Stéphane Robin (MIA, AgroParisTech/INRA).
As an output to this interaction extraction, it was possible to apply a clustering procedure (work of Y. Guédon and J. Legrand), based on a mixture model of random graphs. This allowed us to determine a well-founded simplified network, which was then used to develop a differential equation model. This model showed that the meristem could present well differentiated buffering abilities in its role of auxin perception, a prediction that was corroborated by experiments using a newly developed auxin sensor. The combination of computational techniques and novel experimental tools and data that were developed in this project have led to a new view of the meristem behavior, where auxin signaling, and not only its transport, plays a crucial role. These results have appeared in a systems biology journal with wide readership [16] .
As a follow up, extensions of the differential equation model are under study, with the aim of better understanding this system in more general contexts than the shoot apical meristem development.
-
Complex dynamics and spatial interactions in gene networks (Yassin Refahi, Etienne Farcot, Christophe Godin)
Complex computational and mathematical questions arise in the study of gene networks at two levels: (i) the single cell level, due to complex, nonlinear interactions, (ii) the tissue level, where multiple cells interact through molecular signals and growth, so that even simple local rules can challenge our intuition at higher scales.
At the single cell level, new results were obtained in the framework of piecewise-linear models. Since their introduction in the late 1960's, these models have been believed to present chaotic behavior in some parameter regimes. However, this was mostly observed numerically, based on intensive generation of random networks. In a collaboration between E. Farcot and R. Edwards (Univ. Victoria, Canada), with recent input from one of his students, E. Foxall, we have introduced a method to explicitely build piecewise affine models having a return map which is conjugate to a topological horseshoe. A paper presenting these results is currently in revision.
At a higher scale, we have also continued the study of gene regulation in meristematic tissues. Numerical tools for the simulation of gene network models have been further developed, in the context of Y. Refahi's thesis. Using realistic 3D and 4D structures, reconstructed from microscopy data using the methods in [5] , we have defined putative gene expression patterns in collaboration with biologist colleagues in Lyon (P. Das). We then have constructed an adimensionalized model with only 3 parameters, based on the existing literature [37] . Using standard optimization procedures to fit these parameters we were able to reproduced the in silico patterns with great accuracy. Moreover, alternative patterns were found, based on hypotheses of continuous deformation of gene patterns under tissue growth. These predictions remain to be tested, and this will constitute a large part of Y. Refahi's post-doctoral project (INRA/INRIA CJS fund), in collaboration with ENS Lyon and the team of Henrik Jönsson in Lund, Sweden.
A related study has been initiated during a visit of E. Farcot at the Memorial University of Newfoundland, funded by the INRIA "Explorateur" program (see international relations section). This visit has allowed to initiate a mathematical study of gene regulated patterning in plants, with the aid of tools from equivariant dynamical systems theory, and more generally by relying on symmetry properties. Some first results have been obtained concerning homogeneous patterns in regular tissues, and their bifurcations.
Model integration
Participants : Mikaël Lucas, Michael Walker, Jérôme Chopard, Frédéric Boudon, Christophe Godin, Laurent Laplaze, Jan Traas, François Parcy.
This research theme is supported by the ANR/BBSRC project iSam.
Our approach consists in building a programmable tissue which is able to accept different modeling components. This includes a central data structure representing the tissue in either 2-D or 3-D, which is able to grow in time, models of gene activity and regulation, models of signal exchange (physical and chemical) between cells and models of cell cycle (which includes cell division). For each modeling component, one or several approaches are investigated in depth, possibly at different temporal and spatial scales, using the data available from the partners (imaging, gene networks, and expression patterns). Approaches are compared and assessed on the same data. The objective of each submodel component will be to provide plugin components, corresponding to simplified versions of their models if necessary, that can be injected in the programmable tissue platform.
-
Development of a computer platform for the 'programmable tissue'. (Jérôme Chopard, Michael Walker, Frédéric Boudon, Etienne Farcot, Christophe Godin)
One key aspect of our approach is the development of a computer platform dedicated to programming virtual tissue development. This platform will be used to carry out integration of the different models developed in this research axis. The platform is based on OpenAlea. Partner models can be integrated in the platform in a non-intrusive way (the code of their model need not be rewritten). In this context, model integration will i) consist of designing adequate data-structures at different levels that will be exchanged and reused among the different plug-in models and ii) defining control flows at adequate levels to avoid the burden of excessive interaction between components.
-
Design of a genetic model of inflorescence development. (Etienne Farcot, Christophe Godin, François Parcy)
At a scale involving organs and their geometric arrangement, we have developed a first model of the control of floral initiation by genes, and in particular the situation of cauliflower mutants, in which the meristem fails in making a complete transition to the flower. This work couples models at different scales, since gene regulation is described by a minimal gene network, which is used as a decision module in an L-system model of the inflorescence architecture. This coupled model has led us to hypothesize some interactions between genes and a particular plant hormone, and experiments are currently being made to verify this prediction.